Search
实时荧光定量 PCR (qPCR) 和全转录组 NGS——不需要二选一

实时荧光定量 PCR 和 NGS 的优缺点
随着处理基因表达等应用的新技术和方法的出现,很难确定哪种方法更好,一种方法是否可以简单地代替另一种方法,或者搭配使用能否提供更完美的结果?在此,我们尝试阐明如何在实时荧光定量 PCR (qPCR) 和下一代测序 (NGS) 之间进行选择,或者您是否真的需要择其一。
寻求商业化?立即联系我们的 OEM 服务团队 ›
实时荧光定量 PCR 和全转录组 NGS
全转录组 NGS 有两种主要方法:RNA-Seq 和靶向转录组。当以下一个或多个项目对于您的研究工作至关重要时,建议使用 RNA-Seq:
- 您希望检测所有细胞 RNA:mRNA、miRNA、tRNA 等
- 您希望了解转录本同源异构体的多样性/表达
- 新发现至关重要,即您希望在未先行了解哪些靶标可能受未来治疗影响的情况下鉴别差异表达基因
靶向转录组方法有相似之处;尽管输出将限于较常见的 mRNA 转录本,但在涵盖大多数已知转录组的20,000个靶标范围内,通常可能有新发现。
实时荧光定量 PCR 在哪些方面适合以下工作流程?数据完整性是基因表达实验的关键,因此普遍做法是在 NGS 的上游和下游均使用实时荧光定量 PCR。这意味着,我们并非必须在实时荧光定量 PCR 与 NGS 中择其一;恰恰相反,这两种技术是互补的,共同使用可以生成值得信赖的结果。在 NGS 的上游,通常使用 TaqMan 实时荧光定量 PCR 来检查 NGS 前 cDNA 的完整性。
在 NGS 的下游,实时荧光定量 PCR 仍然是结果验证的必备方法。若不需要进行验证,务必注意实时荧光定量 PCR 是开展涉及在 NGS 筛选过程中发现的转录本靶向组合的后续研究的金标准技术。
下列出版物重点介绍了 Ion AmpliSeq NGS 与 TaqMan 实时荧光定量 PCR 在 cDNA 完整性检查或结果验证方面的共同应用。
阅读我们表明 TaqMan 基因表达测定、Clariom D 测定和 Ion AmpliSeq 转录组试剂盒之间高度一致的技术资料。
采用 Ion AmpliSeq 和 TaqMan 这两种技术的 BIOZ 生物医学研究文章
实时荧光定量 PCR 和 Illumina 靶向扩增子 RNA-Seq
Illumina 靶向扩增子 RNA-Seq 是一种 NGS 方法,可检测感兴趣的 12-1,200 个转录本。此靶标范围与使用实时荧光定量 PCR 可能得到的靶标范围重叠(图1)。在实时荧光定量 PCR 和 Illumina 靶向扩增子 RNA-Seq 之间选择时,应考虑以下因素:
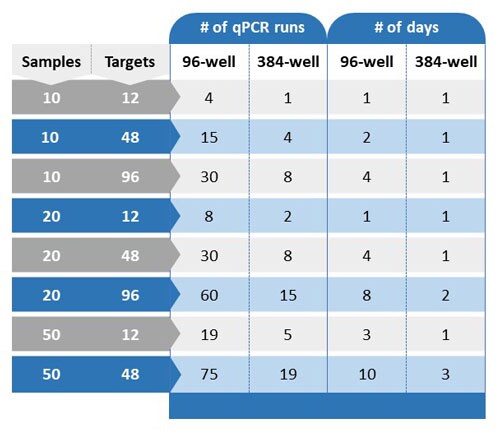
- 过去,实时荧光定量 PCR 一直被视为基因表达定量的金标准。因此,实时荧光定量 PCR 仪器无处不在,实验台和数据分析工作流程非常常见且简单。对于涉及20份样本和10个靶标的实验(冷冻冰箱中已有 RNA 样本和 qPCR 测定试剂),在几张96孔板上运行 1-2 天即可生成所有数据。使用 Illumina 靶向扩增子 RNA-Seq 进行同一实验则需要更长的时间,特别是当无法轻易使用 Illumina 仪器时。将样本发送给核心机构或服务提供商可能会增加从样本到结果的周转时间。
- Illumina 靶向扩增子 RNA-Seq 也将花费更多费用。对于靶标少于20个的实验而言,靶向扩增子 NGS 成本更高。
- 最后,由于 NGS 具有绝对读段计数的特性,因此人们常常认为 Illumina 靶向扩增子 RNA-Seq 的灵敏度和动态范围优于实时荧光定量 PCR。值得注意的是,在大多数实验环境下,实时荧光定量 PCR 的灵敏度和动态范围都足够,而 Illumina 靶向扩增子 RNA-Seq 实现高灵敏度经常需要较大深度的测序,这将增加实验的成本。

常见问题解答
可以,TaqMan 基因表达测定可用于我们预设计的测定系列中的所有基因和种属的大部分外显子-外显子连接。 要选择变体专用的测定,请先确定 NCBI RefSeq 转录本登录号,然后在 TaqMan 测定搜索工具中使用该登录号进行搜索。 在随后出现的测定列表中,选择仅检测感兴趣的那个变体的测定。 如果所有可用的测定均检测超过一个 RefSeq 转录本,则使用我们的定制测定设计工具尝试设计变体专用的测定。
有,TaqMan 阵列板是含有 8-384 种测定试剂(预点样并干燥,您只需添加 cDNA 和预混液)的96孔和384孔板。 根据所需的样本和靶标数量,订购适量的板来实现技术复制和生物学复制。 TaqMan 阵列卡是含有 12-384 种测定试剂的384孔微流控卡,其具有预混液用量比传统板少的附加优势,还具有超简单的台式工作流程:在2分钟内对384个孔进行移液。 TaqMan OpenArray 板是适用于 qPCR 的较高通量选项,每个数据点的价格较低。
事实上这得看情况。 一次 Illumina 靶向扩增子 RNA-Seq 运行可分析的样本和靶标数量取决于 Illumina 平台和化学试剂。 假定所需数量的样本和靶标适合一次 Illumina 靶向扩增子 RNA-Seq 运行,则从样本制备到数据分析的时间为两天。 如果您必须将 Illumina NGS 实验外包给核心机构或服务提供商,可能需要数天或数周才能获得结果。 与之相比,以下实时荧光定量 PCR 实验在 1-3 天内就可完成:
仅供科研使用,不可用于诊断目的。
